Prenomics: Lowering barriers to participation
Posted on 20 October 2022
Prenomics: Lowering barriers to participation

This post first appeared on the Cloud-SPAN forum. Last year, the first Genomics course was delivered, and Community Manager and FAIR coordinator, Evelyn Greeves, blogged about what we learned which included dividing the course into Prenomics and Genomics. Now Evelyn is back with another blog on how that went.
For most of our new learners, the idea of using the command line and accessing the cloud is completely new and a bit overwhelming. But these are skills which, once learned, can totally transform how you do your analysis. That’s why Cloud-SPAN offers a core ‘Prenomics’ course to help you gain confidence in these key skills and prepare you for the Genomics course.
How Prenomics was born
When we ran our first Genomics course in the autumn of 2021, we discovered that a central sticking point for many participants was directory structure and navigation. This was making it really difficult to help people log into the cloud, where we could then teach them core command line skills. We spent much more time troubleshooting and going over these basic skills than we’d expected, meaning we had to rush through more complex content about analysis tools and pipelines later.
Overall, this meant our learners began their command line journey from a place of frustration (which has a huge impact on confidence) and we were forced to minimise the amount of time spent on the ‘genomics’ part of the Genomics course.
As a result, we split the original Genomics curriculum across two courses: Prenomics and Genomics. In Prenomics, all the focus is on the command line. We teach about files and directory structure and spend lots of time guiding learners through creating a folder and navigating to it without using the command line. We also teach those crucial core skills - commonly used commands like ls, cd and mv. Later, as learners gain confidence, we move on to more complex commands like grep and redirection with > and >>.
Why is Prenomics important?
Separating the curriculum into two courses has several benefits. Most importantly, it gives us lots more time to dedicate to core skills and troubleshooting. It also gives us more time to spend on Genomics, as learners start that course already confident about their command-line abilities. Plus, those already familiar with the command line can skip Prenomics and go straight to Genomics.
And it works! We know this because we collect evaluation feedback from participants both before and after the course, asking them to evaluate their understanding of various topics. After we ran the newly designed Prenomics/Genomics combo course in Spring 2022, we compared the understanding scores of learners across three general themes of Command Line, Genomics and Project Organisation (a higher number means a higher level of understanding, with no understanding being 1 and most confident understanding being 6).
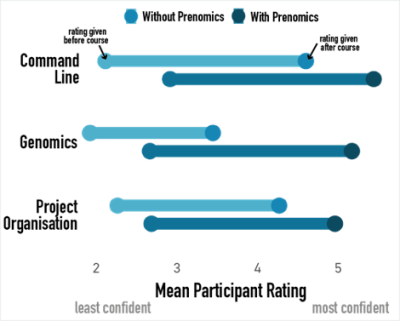
As we’d hoped, we saw improvement in understanding across all themes following both courses. However, when we ran Genomics without the Prenomics prerequisite, we saw a relatively small increase in understanding of Genomics topics. On average, participants finished the course with an understanding level of between 3 (“used a bit but not comfortable”) and 4 (“fairly comfortable in some aspects”).
In comparison, after introducing Prenomics we can see that participants started Genomics with generally higher levels of understanding across all three themes. Even better, looking at the average improvement for each theme shows that participants showed greater improvement in understanding in the Genomics theme when they had Prenomics training beforehand.
This is great news because it means our decision to add the Prenomics module was a good one! We will continue to make tweaks to our courses to benefit our learners and collect feedback to tell us whether they are helpful.
Register Now
If you’re interested in registering for Prenomics, you can do so now.
The next course runs in November 2022 and is completely free, as are all of our courses.
Upcoming Courses
Prenomics – 22-23 November 2022 (online workshop or complete via self-study).
Genomics – 6-7 December 2022 (in-person workshop or complete via self-study).
Genomics self-study – Registration is now open. Start at any time.
About Cloud-SPAN
Cloud-SPAN is a collaboration between SSI Fellow Emma Rand and James Chong (both University of York) and Neil Chue Hong, SSI Director, which aims to upskill bioscience researchers in specialised analyses for Environmental Biotechnology with Cloud-based High-Performance Computing. It is funded by the UKRI innovation scholars award (MR/V038680/1).

