A review of Cloud-SPAN’s Genomics course
Posted on 21 December 2021
A review of Cloud-SPAN’s Genomics course
By Evelyn Greeves.
First appeared on the Cloud-SPAN website.
Cloud-SPAN is collaboration between the SSI fellow Emma Rand and James Chong (both University of York) and Neil Chue Hong, director of SSI which aims to upskill bioscience researchers in the specialised analyses for Environmental Biotechnology with Cloud-based High Performance Computing. It was funded by the UKRI innovation scholars award (MR/V038680/1). At the end of November we delivered our Genomics course for the first time! Our Community Manager and FAIR coordinator, Evelyn Greeves, blogged about what we learned.
Well, it's been a couple of weeks since we ran our first course and we've certainly learned a lot from it! Most of the problems we stumbled into were minor teething issues, but we're also going to be making some fairly major changes based on what we learned. Here's a bit of an update:
First things first... positive feedback!
We had some amazing feedback from our first run-through! We asked our participants to rate their level of comfort with a number of topics before and after completing the course.
As you can see in the graph below, on average our participants felt that their level of comfort had improved after taking the course for all topics. This is great news!
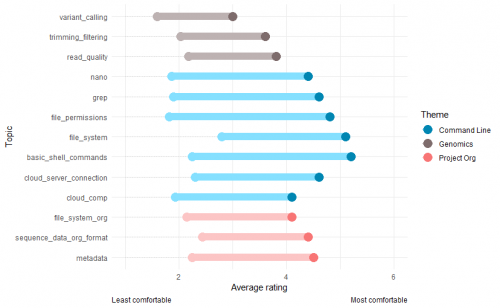
In particular, participants ended the course feeling really comfortable with using the command line to navigate file directories, create and modify files and search for keywords.
The big problem: Time flies
The biggest problem we ran into was time. We just didn't have enough of it and that means we need to seriously rethink how we structure the content of our course. You can see from the graph above that the topics people felt they had improved their confidence the least in were those relating to genomics - assessing read quality, trimming and filtering, and variant calling.
That's because before we could teach those topics, we had to make sure people felt really comfortable using the command line and organising their files. Unfortunately, as we got more and more behind schedule, this meant the genomics topics got less and less time allocated to them.
This is a problem, because the title of the course was Foundational Genomics. If participants are finishing our course feeling like they are not comfortable with variant calling (which is what a score of 3 means) then we need to make some changes. Plus, the genomics bit is meant to be the fun bit!
The written feedback we had from participants after the course, while mostly positive, generally reflected this sentiment too.
The big solution: Course structure
In light of this feedback, we've decided to split our 4 x half-day Foundational Genomics course into two shorter courses: one 2 x half-day "Prenomics" course and one 4 x half-day "Genomics" course.
The Prenomics course will be aimed at complete beginners and will be a gentle introduction to files, directories, relative and absolute paths and basic command line commands. By giving ourselves two half-days to cover this content we should be able to ensure all participants are fully on board before we move onto anything more complex.
This course will also be optional for those who already have some basic experience using the command line. We plan to allow potential attendees to self-screen based on their experience using a series of questions to help them determine whether they would benefit from attending the Prenomics course, or could go straight to Genomics.
The Genomics course will follow directly on from Prenomics, using many of the same commands and building on them further. We should have much more time to cover essential topics such as read quality and variant calling which were rushed last time, allowing us to offer a much better educational experience.
Some smaller problems which this change also solves...
- We plan to run the Genomics course over two weeks, with 2 x half days per week. This, combined with a hopefully much less stressful volume of content to cover, will decrease pressure on our instructors and helpers, most of whom have multiple other commitments.
- Some of our participants felt the course went far too slowly; others felt it went too fast. By screening for those who have the least experience working in the command line, we should be able to both provide more support to those who need it and stop more experienced participants from getting bored.
When will these be delivered?
Prenomics will be delivered from 10:00 - 13:00 on 8th and 9th March 2022.
Genomics will be delivered from 10:00 - 13:00 on 14th, 16th, 21st and 23rd March 2022.
Registration will open on our website soon but to register your interest now please contact cloud-span-project@york.ac.uk.
Other updates in the pipeline
These two new courses, Prenomics and Genomics, will make up the 'Foundational' element of our content. We still have plans to develop 'Advanced' modules on topics such as automation and setting up cloud instances. We asked our participants to let us know what they'd like to see, and we'll be taking these suggestions into account as we go.
We also have plans to run some (hopefully) in-person 'hack days' for alumni of our Foundational courses, where we can help participants apply the skills learned in the course to their own problems, and assist in troubleshooting any problems. We hope this will help develop our community of practice further by providing opportunities for networking and building relationships.
And that's it. Thanks for reading this far - we hope it explains some of our rationale for why the course is structured the way it is. We're really proud of our achievements with our first ever course, and we're looking forward to trying some new stuff out next time.

