PickCells and exploratory image analysis in cell biology
Posted on 13 September 2018
PickCells and exploratory image analysis in cell biology
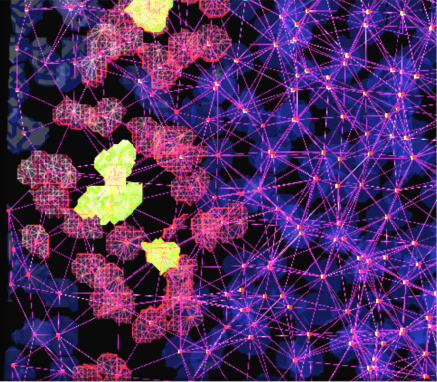 Image courtesy of Sally Lowell
Image courtesy of Sally LowellBy Mike Jackson, Software Architect, The Software Sustainability Institute.
PickCells is an image analysis platform developed by the Centre for Regenerative Medicine (CRM) at The University of Edinburgh. PickCells combines generic image analysis algorithms, visualisation modules and data mining functionality within a stand-alone Java application. PickCells provides a graphical environment within which biologists can study multidimensional biological images and explore 3D spatial relationships between objects within complex biological systems such as stem cell niches, organoids, and embryos. Since January, EPCC has been working with CRM on the development of PickCells and its supporting resources.
My EPCC colleagues Elena Breitmoser and Arno Proeme and myself worked with CRM's Sally Lowell and Guillaume Blin to take PickCells into a state suitable for more widespread promotion with the intent of encouraging deeper community engagement, by both users and developers. Our work was focused on building and populating a web site for users, developers and contributors and providing consultancy on developing and supporting open source software.
PickCells is highly-modular and the documentation for each component is held within the associated source code repository for that component. These source code repositories are hosted by Framagit, a deployment of GitLab for free open source software. A web site framework was developed using the Hugo static web site generator to render content and GitLab's continuous integration functionality, GitLab CI, is used to trigger rebuilds of the web site in response to any changes in documentation held within any of the source code repositories. Though it is still being populated, you are welcome to browse the evolving PickCells web site.
Our work was funded by the Wellcome Trust Institutional Strategic Support Fund and The Software Sustainability Institute.
During our collaboration, Sally secured a Wellcome Trust "enrichment" award to support further collaboration on PickCells. This next phase of work, which begins in September, seeks to improve the usability of PickCells, from the perspective of biologists who wish to extract information from their imaging data. Our objectives are to develop in-application interactive help to help users get started with analyses using PickCells; filter information (e.g. displays, menus, or icons) depending upon workflows being followed by users; produce video tutorials on how to use PickCells; undertake a usability evaluation of PickCells, using researchers at CRM as evaluators and soliciting feedback from the wider PickCells community; and to update PickCells and its supporting resources based upon the outcomes of this usability evaluation. We look forward to reporting on our experiences in a future post.
