Speeding up Python with Cython—some thoughts for Python programmers
Posted on 25 July 2017
Speeding up Python with Cython—some thoughts for Python programmers
 By Thomas Etherington, Senior Research Leader, Royal Botanic Gardens, Kew, and Software Sustainability Institute Fellow.
By Thomas Etherington, Senior Research Leader, Royal Botanic Gardens, Kew, and Software Sustainability Institute Fellow.
Getting code running fast enough to be useful is an important consideration for making software sustainable. For Python programmers, the Cython project provides an opportunity to speed up your Python code. As part of my Software Sustainability Institute Fellowship, I spent a couple of days learning about Cython from one of the lead developers, and I’ve summarised from my perspective when Cython could be a useful tool for others to explore.
My interest in Cython began when I looked into the code of a SciPy function and saw code that looked quite Pythonic, but clearly wasn’t actual Python code. It transpires that a lot of SciPy functions have been written using Cython, which is a language that can either: compile Python code directly to C, or wrap C or C++ code in Python, so that computational speeds associated with lower-level C programming can be leveraged from a higher-level Python programming interface. So while SciPy is one of my favourite Python packages, the code itself actually consists of “more than 200,000 lines of C++, 60,000 lines of C, [...] compared to about 70,000 lines of Python code” (Behnel et al. 2011, p.31). Therefore, I wanted to learn more about Cython in the hope of being able to contribute to the future development of SciPy, and possibly discover ways to write better code of my own. After a couple of failed self-learning attempts, I recently spent two days at the Python Academy in Leipzig learning about Cython from one of its lead developers, and this blog summarises my main thoughts about Cython as a potential tool for Python programmers.
Let’s start with an example that shows why you might want to consider using Cython. Travis Oliphant has written a blog post that demonstrates nicely how with a few extra lines of code and some explicit type declarations, Cython can produce results nearly 600 times quicker than the original pure Python approach! This seems quite magical, so it perhaps begs the question: why aren’t we all using Cython?
I think the first answer to that is also in Travis Oliphant’s blog. While Cython was nearly 600 times faster than pure Python, it was only just over twice as fast as an equivalent NumPy approach. The reason that NumPy is so fast is that NumPy uses vectorised operations that are actually implemented in C (van der Walt et al. 2011). So like SciPy, while NumPy looks to be written Python, it is in fact leverage the power of C. For a Python programmer like myself who doesn’t know C, it is perhaps no surprise that given I can easily write Python code with NumPy to get near C speeds. This will perhaps suffice for my speed requirements, and hence, I may not even need to consider another option like Cython.
A second answer I think relates to the need for Cython to compile code. As someone who only programs in interpreted languages such as Python and R, the concept of compiling code was a new one to me. Perhaps the key point to understand is that compiled code is platform dependent. This means that your code needs to be either compiled by another potential user, which might limit the potential audience as only those capable of compiling code, or provided and maintained for a variety of platforms, which creates additional work for a developer.
So, having learnt more about Cython, will I be using it? I think it’s important to remember that every use case will be different, so it’s hard to provide specific rules that are widely applicable. However, I suspect that I can usually achieve the computational speeds I need using NumPy, and that any potential Cython speeds gains would probably be outweighed by the additional time I would need to develop and distribute my code—which is likely to be considerable given my lack of C skills! However, there may well be occasions when I do need something faster than NumPy, or there may be code available in C for something that is not available in NumPy. In such a situation Cython gives me another tool that I can turn to. Also, even if I don’t develop the code myself, I now understand Cython sufficiently that I am now in a position where I could collaborate with a Cython or C programmer—which may well be a more efficient approach! So, assuming there are other people like me out there, who program solely with Python, I propose the following decision-tree to try and help them decide when Cython might be something to explore further themselves:
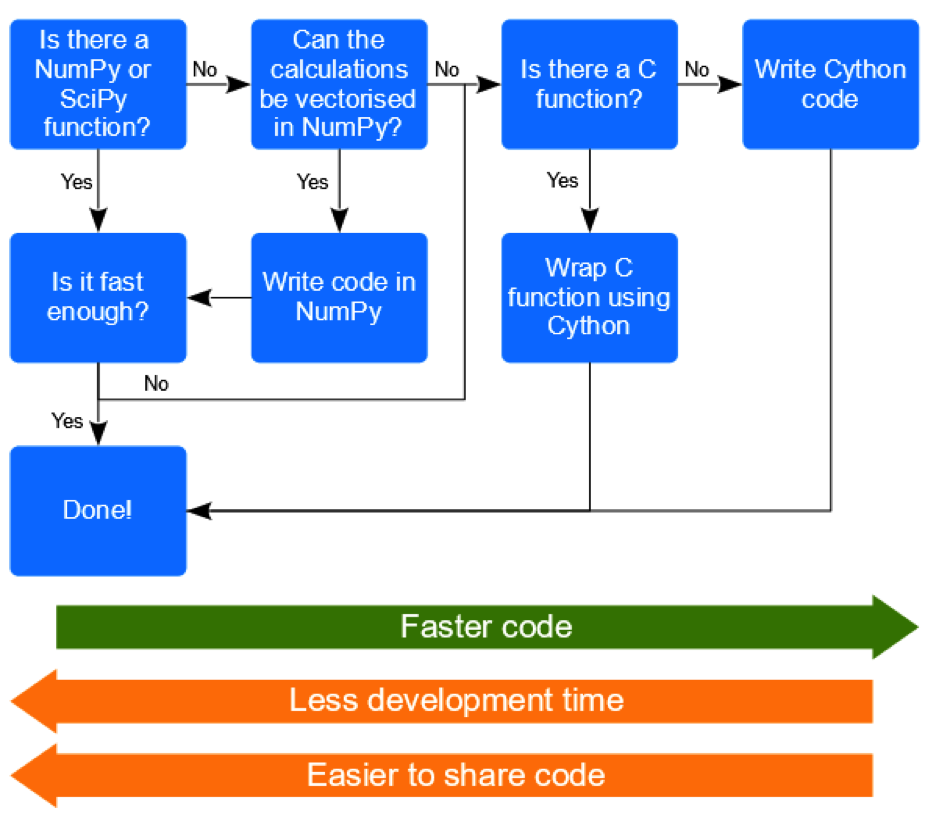
For those Python programmers who do want to explore Cython further there are some useful starting points. I thought the Cython course I attended was quite good, and I particularly liked that it was taught by one of the lead developers of the project. So for people with a decent level of Python programming experience this could be a good choice, and while not essential, I think I would’ve been able to get to grips with things better if I had some basic C knowledge. But if you can’t get to a course, then the Cython project tutorials may be a good option. There are some openly available SciPy conference papers (Behnel et al. 2009, Seljebotn 2009) that will provide some more detail about what Cython can do and how to go about doing it.

